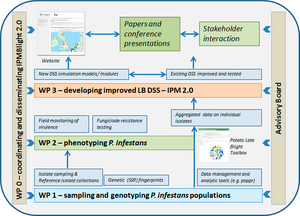
WP descriptions
The foundation for this project is recommendations from the EuroBligt workshop in Brasov, May 2015. The work programme is organised according to the work packages (WP) described below, with the inter-relationship expressed in the project diagram given at the bottom of this page.
WP0. Coordinating and disseminating IPMBlight2.0 activities | ||
Didier Andrivon, INRA. Lead of WP0 | Overall project management and responsibility will be carried out by the project coordinator, Didier Andrivon, and the Project Management Board composed of the project coordinator and Workpackage (WP) leaders. Advisory Board. We will set-up an Advisory Board (AB) comprising one expert from each sector: science, extension, breeding and the chemical industry. All AB members will be external to the project, so as to provide independent advice and assessments. Internal communication & IPR and contractual issues. A Consortium Agreement will be prepared and signed in the first month of the project, to secure an open and transparent process about research agenda, use of data and IPR related issues. | |
WP1. Monitoring European P. infestans populations. | ||
David Cooke, JHI. Lead of WP1 | WP1 complements the survey undertaken by EuroBlight, and will enable Europe to track emerging populations. The work will build on existing achievements by the EuroBlight network and associated global partners with regard to monitoring of P. infestans in Europe and beyond. The WP work will answer questions like: What are the dominant clonal lineages (genotypes) in Europe over time and across countries? What is their relatedness to global populations? How can we quickly and reliably identify new emerging populations? This WP will build the foundations for a global service enabling Europe to track and trace new emerging populations originating from Europe itself or from other continents. | |
WP2. Linking phenotypes to genotypes | ||
Eve Runno-Pauerson, EULS Lead of WP2 | This WP aims to analyse key phenotypic traits (aggressiveness – i.e. quantitative pathogenicity; sensitivity to major active ingredients currently in use in Europe for late blight control; virulence – i.e. ability to infect breeding material or in European potato cultivars possessing race-specific resistance genes) from the isolate collections established and genotyped in WP 1. The comparison of genotypic and phenotypic data will be carried out to determine the possibilities of inferring phenotypic characteristics from genotypic markers, in both clonal and sexual populations of the pathogen. If possible, this inference will be very useful for prediction and strategy design purposes, as the genotypic fingerprinting is much quicker than phenotypic testing. We will therefore analyse variation both between genotypes and within clonal lineages, as these lineages often consist of numerous variants. | |
WP3. Developing innovative DSS integrating epidemiological and pathogen population knowledge | ||
Jens Grønbech Hansen Lead of WP3 | The objective is to develop a new, quantitative approach to integrate knowledge on weather-based infection risk, epidemiological parameters, and pathogen phenotype and genotype characteristics (IPM2.0 concept). A suite of new on-line simulation models, making use of the phenotypic and genotypic data, weather data, and trap nurseries data from WP1 and WP2, will be designed and implemented as web-based tools. Existing DSSs in partner countries will then be adjusted based on results from step 1, and improved DSSs will be tested in field experimental trials. | |