Potato blight tracking in Europe
A team of researchers examining the ongoing evolution of the European population of the potato late blight pathogen now report on the 2016 data. Over 1600 samples were genotyped from 26 countries and entered into the EuroBlight database. More than 70% of the samples were of defined clonal lineages that have been observed in previous seasons. Some clones are widespread and have been present in Europe for more than a decade but two recently emerged clones showed marked local expansion in the 2016 season. The other 30% of the population comprised ephemeral genetically diverse isolates consistent with a source of oospore-borne inoculum. A clear regional pattern in the dominance of clones versus sexual recombinants was observed across Europe.
Since its arrival in the nineteenth century, Phytophthora infestans, the cause of late blight has remained a serious threat to potato crops. Although we are now better equipped to control the disease than in the past, evolving pathogen populations continue to challenge our management practices. Rapid changes in P. infestans populations causing late blight in Europe, America and Asia, including the emergence of strains with increased aggressiveness or reduced fungicide sensitivity have been observed. We have been monitoring populations and characterising the invasive genotypes to help optimise IPM strategies, as required by EU Directive 2009/128/EC on the sustainable use of plant protection products. The changes in P. infestans populations directly influence the development and deployment of resistant cultivars, the performance of disease warning systems and the efficacy of plant protection products. Coordinated and continuous pathogen monitoring was proposed by the EuroBlight consortium at its meeting in 2013.
As in previous years, ‘FTA cards’ were distributed to disease ‘scouts’ from across the industry who visited blight-infected crops. Disease lesions were pressed on the cards and returned to the laboratories where the pathogen DNA was fingerprinted at the James Hutton Institute and INRA Rennes. The DNA fingerprint data defines the clonal lineages of the pathogen and is combined with geo-location data to plot the diversity across Europe. In many parts of Europe, disease pressure was generally high and early in 2016, which resulted in a higher than average sample density and distribution. Over 1600 samples were thus collected by 20 partners across 26 European countries. As in previous years, the AHDB Potatoes ‘Fight Against Blight’ campaign data from British crops is included. This year also includes data from partners in the IPMBlight2.0 project, which is also generating pathogen phenotype data to support IPM strategies. The data from 2013-2016 now comprise over 5250 samples from 34 countries. With the support of international groups, data is also entered for parts of Asia and North Africa.
 |  |
|---|---|
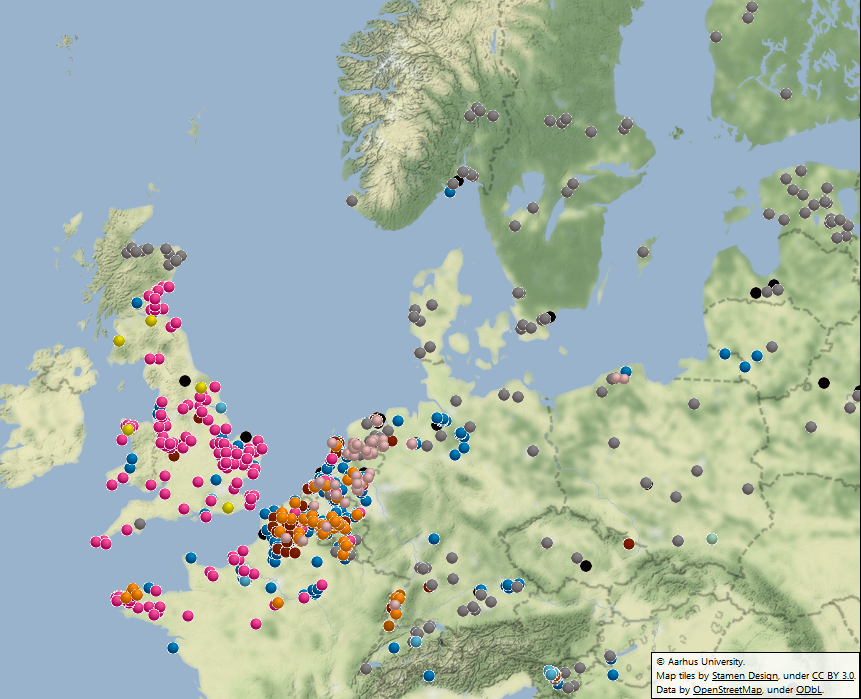
Figure 1. Genotype map, 2016 sampling. Go to live map. |
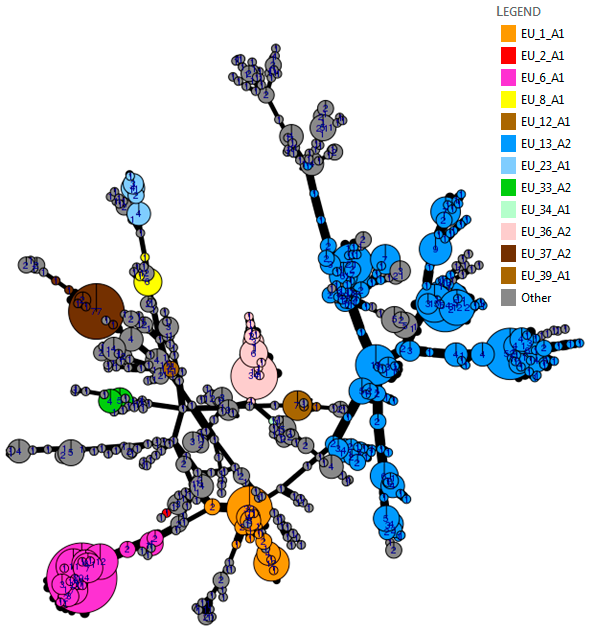
In each of the last four years, 60 to 79% of the population comprised known clonal lineages that recur each season. The remaining samples were novel, genetically diverse genotypes found at a single location in one season and grouped in a category termed ‘Other’ (Figure 1). In 2016, clone EU_13_A2 (blue-13) made up 31% of the samples and was present in 13 countries. The spread of EU_13_A2 and its resistance to metalaxyl is affecting management efficacy in parts of Asia and North Africa, and reinforces the need for pathogen data to support IPM best practice. The second most prevalent clone, EU_6_A1, was found at a 24% frequency in 2016 but was localised to seven countries in northwest Europe.
The frequency of the clone EU_1_A1 decreased from 7 to 4.5% of the population, and was restricted to Belgium, France and the Netherlands. The spread of two new clones is cause for concern. EU_36_A2 was first sampled in the starch potato areas in Germany and the Netherlands in 2014 and again at a low frequency in the low blight pressure 2015 season. In 2016, it increased to almost 4% of the population and spread across the Netherlands into Belgium and northern France. A single sample of EU_37_A2 was first detected in Noordoostpolder in the Netherlands in 2013 and was sampled locally at a low frequency in the following two seasons. However, it comprised 5.5% of the population sampled in 2016, having spread as widely as England, Poland and Slovenia with a cluster of samples in northern France. Their survival and spread at a time when other clones (e.g. EU_34_A1 and EU_35_A2) have failed to establish suggests they are evolutionarily fit and may pose challenges to disease control. A strong ‘founder effect’ in which inoculum of these genotypes spread from the first outbreaks during the highly conducive conditions early in the 2016 growing season provides an alternative explanation. Lastly, the genetically diverse ‘Other’ samples comprised 28% of the population and remained at the highest frequency in the east and north-east of Europe. In contrast to some clones, the traits possessed by the ‘Other’ isolates and the new clones (EU_36_A2 and EU_37_A2) are not yet known. However, the IPMBlight2.0 project (http://euroblight.net/research-projects/ipmblight20) has started a detailed study of the traits of clonal and sexually recombinant isolates to help improve late blight management and forecasting.
The genetic diversity of the 2016 population is visualised using an analysis tool (poppr 2.0) linked to the pathogen database. The minimum spanning network (top chart) visualises the 2016 population structure. Considerable sub-clonal diversity within the EU_13_A2 lineage, which is 12 years old, is observed compared to the 3-4 year old EU_36_A2 and EU_37_A2 clones. The grey ‘Other’ isolates are genetically diverse and distributed across the whole network. More detailed analysis is underway to examine population change using these powerful tools.
This model of pathogen tracking offers a rapid, cost-effective and co-ordinated approach to understanding pathogen evolution on a European scale. Data on the dominant clones has been passed to growers, advisors, breeders and agrochemical companies to provide practical management advice and shape longer-term strategies. Furthermore, early warning of the incidence and spread of recently evolved clones will enable a timely response from the industry.
The Euroblight network is continuing to harmonise methods with other networks in the Americas and Asia and encourages continued co-operation between groups involved in managing late blight to exploit the database and tools for improved awareness and blight management on a global scale. We will continue the project in 2017, so please make contact with the project team if you would like more information. We thank all the partners who have contributed to the funding and data collection.
Companies and institutions that participated in the sampling and sponsored the project (2013-2016)
ADAMA, AFBI, Agrifirm, Agriphar, AHDB Potatoes, ARVALIS-Institut du Végétal, BASF SE, Bayer CropScience AG, Bayerische Landesanstalt für Landwirtschaft, Belchim Crop Protection, Centre Wallon de Recherches Agronomiques, Certis, Cheminova, CropSolutions, Dupont de Nemours, Emsland Group, Estonian University of Life Sciences, Germicopa SAS, HZPC Holland B.V., Neiker, Nordisk Alkali, NIBIO, PCA, The Plant Breeding and Acclimatization Institute (IHAR), Profytodsd, Swedish University of Agricultural Sciences and Syngenta Agro GmbH.
Contacts
- Jens G. Hansen & Poul Lassen, Aarhus University. Contact: jensg.hansen@agro.au.dk
- Geert Kessel, Wageningen University and Research Centre. Contact: geert.kessel@wur.nl
- David Cooke: James Hutton Institute, Dundee. Contact: david.cooke@hutton.ac.uk
- Didier Andrivon, Roselyne Corbieres: INRA Rennes. Contact: didier.andrivon@.inra.fr
Download the news story as pdf here
How to participate? Sampling protocol and sampling form is updated here
 |  |