Results of the EuroBlight potato late blight monitoring in 2021
EuroBlight is continuously examining the ongoing evolution of the European population of the potato late blight pathogen and now reports on the 2021 results. Approximately 2500 samples from 26 countries genotyped.
Key findings:
-
The blight pressure in 2021 was higher than average across many parts of Europe.
-
Disease outbreaks from 26 countries were sampled by several groups in 2021 resulting 2492 genotyped samples.
-
Around 72% of these samples were of defined clonal lineages observed in previous seasons while the rest consisted of ephemeral, genetically diverse ‘Other’ types consistent with oospore-borne inoculum.
-
The proportions of the main clones in the 2021 population were broadly similar to those reported in 2020. Newer clones EU_41_A2, EU_37_A2, EU_36_A2, EU_43_A1, EU_44_A2 and EU_45_A2 made up 48% of the 2021 population. These clones are displacing the established 13_A2, 6_A1 and 1_A1 clones which caused only 17% of the blight lesions sampled in 2021.
-
Regional trends in the type and proportion of clones compared to sexual recombinant populations were observed across Europe. For example, ‘Other’ genotypes more prevalent in Denmark, Norway, Sweden and Finland than across countries to the west of Europe.
-
Some implications of these displacements and ongoing changes are discussed
How did we do it?
Since its arrival in the nineteenth century, Phytophthora infestans, the cause of potato late blight, has remained a serious threat to European potato production. Although we are now better equipped to control the disease than in the past, evolving pathogen populations challenge integrated management practices. Changes in P. infestans populations directly influence the deployment of resistant cultivars, the performance of disease warning systems and the efficacy of plant protection products.
Co-ordinated and continuous pathogen monitoring was thus proposed by the EuroBlight consortium at its meeting in 2013 and has since been implemented as an EU-wide monitoring activity supported by many stakeholders. Such monitoring and characterisation of the population changes and invasive genotypes helps optimise IPM strategies, as required by EU Directive 2009/128/EC on the sustainable use of plant protection products.
FTA sampling cards onto which lesions had been pressed were returned to laboratories at the James Hutton Institute, Dundee and INRAE, Rennes for pathogen DNA fingerprinting using simple sequence repeat markers. Samples were assigned to existing clonal lineages or defined as new genotypes and all results uploaded to the EuroBlight database. Support from international groups is generating similar data for parts of Asia, South America and Africa, allowing for a more global understanding of pathogen population changes.
The genotype data from 2013-2021 now comprises over 15,000 samples from 37 European countries.
What did we find?
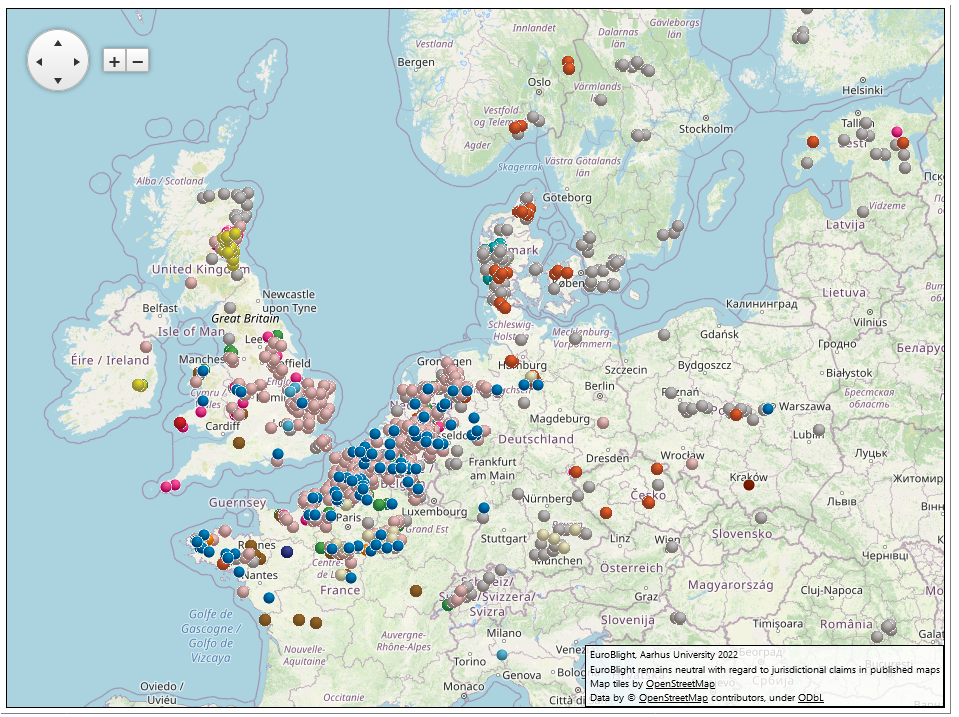
The decline in the clonal lineage EU_13_A2 (blue-13) continued in 2021 as its frequency reduced to 5% of the samples. However, this metalaxyl-resistant clone continues to affect management efficacy in Europe, parts of Asia and North Africa, reinforcing the need for pathogen data to support IPM best practices. The frequency of EU_6_A1 also declined to 12% and was only sampled in France, the UK and Ireland (Figure 1). EU_1_A1 is now almost extinct. Overall, the frequency of these three previously dominant lineages has fallen from almost 68% in 2014 to just over 17.5% in 2021. Over the same time the newer clones have increased in frequency from less than 1% in 2014 to 50% in 2021.
Clone EU_36_A2, first sampled at a low frequency in the starch potato areas in Germany and the Netherlands in 2014, comprised nearly 37% of the 2021 samples and has further established as the dominant clone sampled in NW Europe. Clone EU_41_A2 which was first recorded in Denmark in 2013 reduced in overall proportion but expanded its range to the west into Belgium, The Netherlands, UK, France and Iceland in 2021. A genotype that made up 22.8% of the 2020 Danish samples was defined as EU_43_A1 and also spread west to The Netherlands and Belgium. Two clones were newly defined in 2021, EU_44_A1 and EU_45_A1. The former clone was not reported prior to 2021 but then found across five countries (UK, IE, CH, BE and FR) in a single year. EU_45_A1 first reported in Germany in 2019 spread to France in 2021.
Figure 1. Genotype map - 2021. Go to live map.
The overall frequency of clone EU_37_A2 has been stable at 7 to 8% over the past three years down from its peak of 14% sampled in 2017 and 2018. The reduced sensitivity of EU_37_A2 isolates to fluazinam reported by Wageningen University in 2017 confers a selective advantage to this clone when this product is applied, and probably explained its increasing prevalence in Europe over the 2016 to 2018 seasons. The change in product use and the population response suggests management failures have been avoided by using more effective chemistry and indicates a strong positive benefit of the EuroBlight monitoring approach.
The survival and spread of these newer clones, when others are decreasing or have failed to establish, suggests they are evolutionarily fit and supports anecdotal evidence that they are more challenging to manage in the field. Ongoing fungicide sensitivity testing with a range of key active ingredients in The James Hutton Institute do not however, indicate any insensitivity issues with fungicide active ingredients other than fluazinam and phenylamides (discussed above). Further work in controlled field trials is required to identify the factors that drive population displacements.
Lastly, the genetically diverse ‘Other’ samples comprised around 30% of the sampled population in 2021, a frequency that has remained stable since 2014. These diverse genotypes remain most prevalent in crops in the north and east of Europe and are consistent with a soil-borne source of oospores. Although there are epidemiological threats of earlier primary inoculum and theoretical evolutionary advantages to sexual recombination generating new pathogen phenotypes, we do not yet fully understand the practical threats posed by ‘Other’ strains of the pathogen compared to the clones.
The genetic diversity of the 2021 population has been visualised (Figure 2) using an analysis tool (poppr 2.0) linked to the EuroBlight pathogen database. The minimum spanning network shows sub-clonal diversity within each of the clonal lineages. The clonal and within-clone variation is being used to track the evolution and spread of these pathogen populations across Europe and beyond. ‘Other’ isolates (not shown) are genetically diverse and distributed across the whole network.
Figure 2. Minimum spanning tree of 2021 samples.
The EuroBlight model of pathogen tracking is a rapid, cost-effective and co-ordinated approach to understanding pathogen evolution on a European scale. Data on the dominant clones have been passed to growers, advisors, breeders and agrochemical companies to provide practical management advice and help shape longer-term strategies. The data provide an early warning of the incidence and spread of novel clones and the case of EU_37_A2 demonstrates such a timely response.
The EuroBlight network continues to harmonise methods with other networks in the Americas, Asia and Africa and encourages continued co-operation between groups involved in managing late blight to exploit the database and tools for improved awareness and blight management on a global scale. We will continue the monitoring in 2022, so please contact the project team if you would like more information or if you would like to contribute. We thank all the partners who have contributed samples and supported the project.
Thanks to the following participants/sponsors that have contributed to the 2013-2021 monitoring
Aarhus University, ACVNPT, ADAMA, AFBI, Agrifirm, Agricultural Institute of Slovenia, Agrico, Agriphar, Agroscope, AHDB Potatoes, ARVALIS-Institut du Végétal, BASF SE, Bayer AG CropScience Division, Bayerische Landesanstalt für Landwirtschaft, Belchim Crop Protection, BSV Network (France), Centre Wallon de Recherches Agronomiques, Certis, Cheminova, Corteva, CropSolutions, CUConsulting, CZAV, Delphi, Emsland Group, Estonian University of Life Sciences, Eurofins, Germicopa/ Florimond Deprez, Hochschule Osnabrück, HZPC Holland B.V., INRAE, Institute of Plant Protection and Environment in Serbia, The James Hutton Institute, Julius Kuehn Institute, LfL Bayern, Meijer Potato, Neiker, Nordisk Alkali, NIBIO, PCA, The Plant Breeding and Acclimatization Institute (IHAR), Profytodsd, RML Iceland, Swedish University of Agricultural Sciences, Solynta, Agro GmbH, SynTech Research, , Staphyt, Syngenta, TEAGASC, Technical University of Munich, UPL, Van Iperen, Vertify university of Bologna, University of Freiburg, and Wageningen Research.
Contacts
-
Jens G. Hansen & Poul Lassen, Aarhus University. Contact: jensg.hansen@agro.au.dk
-
David Cooke & Alison Lees: James Hutton Institute, Dundee. Contact: david.cooke@hutton.ac.uk
-
Geert Kessel, Wageningen University and Research Centre. Contact: geert.kessel@wur.nl
-
Didier Andrivon & Roselyne Corbiere, INRAE. Contact: Didier.Andrivon@inrae.fr
Download the news story as pdf
 |