Concepts and methodologies
INTRODUCTION
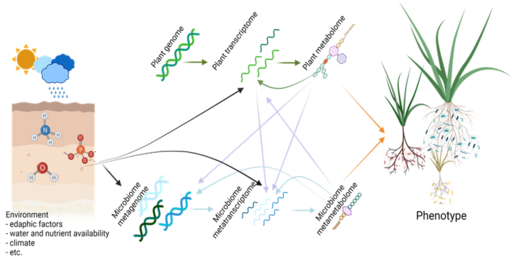
We regard the plant and its associated microbiome as an entity, the plant-microbiome holobiont, where root phenotypic responses are products of intimate interactions between the plant genome and the extremely diverse soil biota.
The composition of the root microbiome is key to its effects on the host. Vice versa, plants, through root exudation of metabolites, selectively recruit soil microbiota that may benefit plant resilience to abiotic and biotic stress.
The greater the belowground diversity, the better the prospects of plant roots of recruiting beneficial microbes to mobilize nutrients, reduce stresses and suppress pathogens. Therefore, the rhizosphere can be considered an extended root phenotype, a manifestation of the effects of plant genes on their environment inside or outside of the organism.
METHODOLOGY
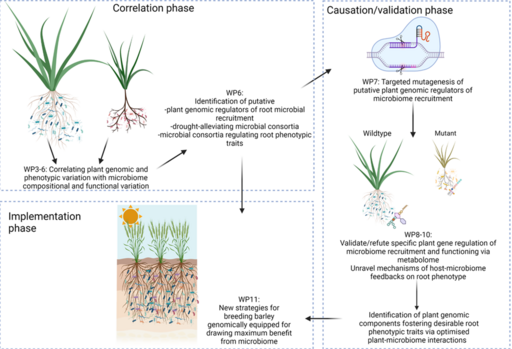
Correlation phase (WP3-6) With a hologenomic approach, we will identify barley genes that are putative regulators of microbiome recruitment and root phenotypic traits: At three field trial locations representing contrasting drought regimes (Morocco, Lebanon and Austria) (WP3), we will grow three diversity panels of 200 barley genotypes each. We will screen root phenotypic traits and root microbiome composition (WP5) of each barley genotype. WP6 will integrate the data on microbiome composition and phenotypic traits from field screenings with barley whole genome sequences (WP4), using GWAS/MGWAS to identify correlations between barley genes, microbiota and root phenotypes and drought resilience.
Causation/validation phase (WP7-10) We will rigorously test causal, mechanistic links between barley genes, microbiome composition/ functioning and root phenotypic drought responses. To do so, WP7 will construct barley lines carrying mutations in genes of interest using TALEN and CRISPR. Under varying drought conditions, we will compare mutants and wildtype lines to verify/refute causal relations between barley genomic regions, the metabolome (WP9), the microbiome composition/functioning (WP10) and root phenotypic traits (WP8).
Implementation phase In WP11, we will exploit the knowledge on barley genetic regulation of root trait plasticity and microbiome composition/ functioning generated in WP3-10 to create strategies for breeding barley varieties fit to tackle drought challenges through synergistic interactions with microbiomes. We will select a subset of barley varieties carrying genomic regions associated with specific root traits and microbiomes from the 600 varieties screened in WP3. These lines will be crossed with elite European lines using speed breeding, and the resulting new varieties will be multiplied and tested against the parental lines.