Results of the EuroBlight potato late blight monitoring in 2019
EuroBlight is continuously examining the ongoing evolution of the European population of the potato late blight pathogen and now reports on the 2019 results. Approximately 1800 samples from 27 countries genotyped.

Key findings:
Three emerging clones of P. infestans (37_A2, 36_A2 and 41_A2) increased their combined frequency from 10% in 2016 to 40% of the population in 2019.
Regional differences exists in the frequency of the emerging clones e.g. the frequency of clone 37_A2 has declined to 10% or less between 2018 and 2019 in the Netherlands and Britain, but remains 25% in Belgium and France.
These recent clones are displacing the established 13_A2, 6_A1 and 1_A1 clones which reduced in frequency from 60 to 30% of the samples collected between 2016 and 2019
A quarter of the population comprised ephemeral, genetically diverse isolates consistent with oospore-borne inoculum.
A regional pattern in the dominance of clones versus sexual recombinants was observed across Europe. Some implications of these displacements and ongoing changes are discussed.
The weather in 2019, was very dry and unfavourable for late blight development in some regions but very conducive for blight in other regions like Denmark and Northern Britain.
Download the extended version of the news story as pdf.
How did we do it?
We ran a EuroBlight coordinated sampling campaign involving the industry and many research institute partners, and analysed the sampled isolates using SSR genotyping at the James Hutton Institute and INRAE as in previous years.
Despite the overall low disease pressure in 2019, 17 partner organisations collected over 1900 FTA card samples from 27 European countries, from which 1816 generated genotype data. The genotype data from 2013-2019 now comprises over 10000 samples from 35 European countries.
Over the last seven years, 60-79% of the sampled population comprised known clonal lineages that recur over multiple seasons. The remaining samples were novel, genetically diverse genotypes found at a single location in one season and grouped in a category termed ‘Other’ (Fig 1).
What did we discover?
For the first time since 2013, the clonal lineage EU_13_A2 (blue-13) was not the most frequently sampled and dropped to 9.3% of the samples collected. Although in decline, this metalaxyl-resistant clone is widespread and continues to affect disease management efficacy in Europe, parts of Asia and North Africa, reinforcing the need for pathogen data to support IPM best practices. The frequency of EU_6_A1 rose to 20.4% due to severe outbreaks in parts of Britain where it remains dominant. The frequency of EU_1_A1 further decreased from 1.6 to 0.4% of the population. A progressive displacement of these three lineages is occurring (Fig 1)
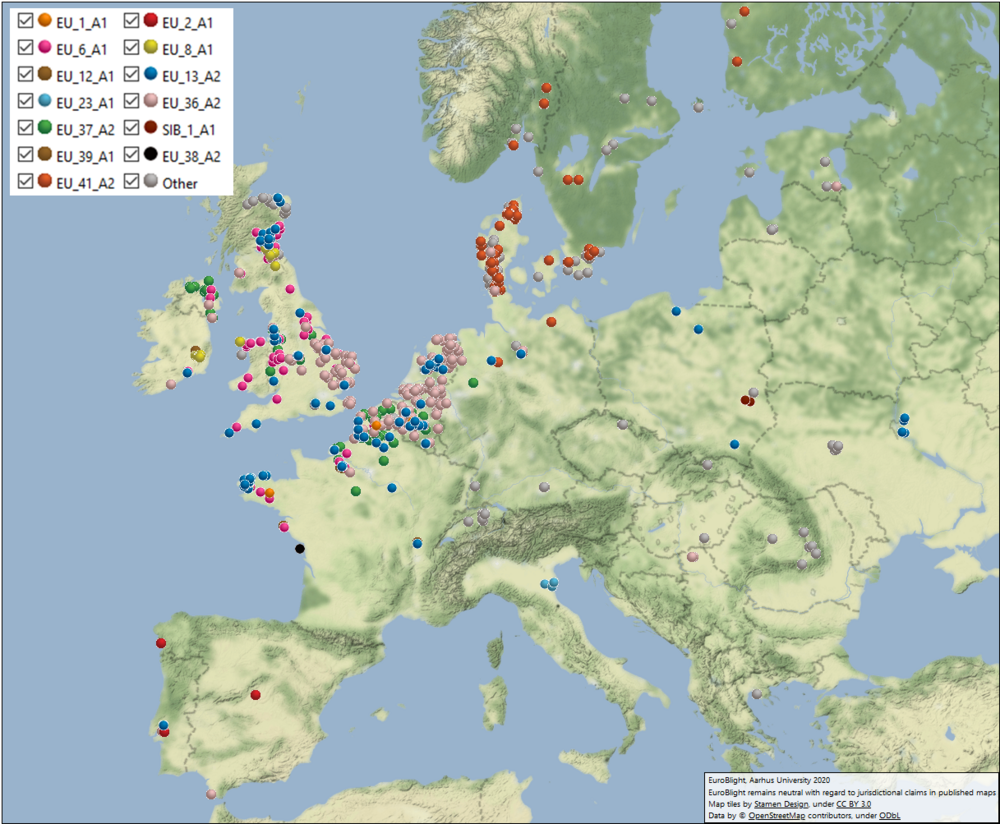
Figure 1. Genotype map - 2019. Go to live map.
Tip!
You can see the same data as frequency pie charts per country on the Genotype Frequency Map and, you can find a chart indicating the evolution across years on the Genotype Frequency Chart. Please note biased data as Britain sampled more data than other countries and some countries have only a few samples analysed (the number of samples is available under the pie charts on the Genotype Frequency Map). On the interactive maps and charts you can also select one or more countries to analyse and you can select one or more years for the generation of summary statistics on maps and charts.
Despite the low blight pressure across many areas, two of the newer clones (EU_36_A2 and EU_41_A2) again increased in frequency in 2019. Clone EU_36_A2, which was first sampled at a low frequency in the starch potato areas in Germany and the Netherlands in 2014, has spread rapidly over the last five seasons. In 2019, it comprised 26% of the European pathogen population, and it was distributed across 15 countries but with a very low incidence in Northeast Europe. Clone EU_41_A2, first recorded in Denmark in 2013, has now spread to five neighbouring countries but has not yet spread west to the low countries. In 2019, its frequency rose from 4.6 to 5.7% of the European population, but now covers 35% of the Danish population. The survival and spread of these clones, when others are decreasing or have failed to establish, suggests they are evolutionarily fit and supports anecdotal evidence that they are more challenging to manage in the field. Conversely, the overall frequency of clone EU_37_A2 declined for the first time since its detection in 2013. It peaked at 14.4% of the sampled population in 2018 but dropped to 8.4% in 2019. The reduced sensitivity of isolates of EU_37_A2 to fluazinam first reported by Wageningen University in 2017 confers a selective advantage to this clone when this product is applied, and probably explains its increasing prevalence in Europe over the 2016 to 2018 seasons. However, widespread reporting of the reduced sensitivity has led to a marked decline in fluazinam use across many countries and the removal of this positive selection pressure possibly explains the overall drop in EU_37_A2 frequency in 2019. This change in product use and the population response suggests management failures have been avoided by using more effective chemistry and indicates a strong positive benefit of the EuroBlight monitoring approach. Regional differences, which may relate to current patterns of fluazinam use, are apparent with EU_37_A2 still comprising 25% of samples in France and Belgium in 2019 compared to falls to 4% and 10% in the Netherlands and England, respectively.
Fungicide sensitivity testing in laboratories in Wageningen University and The James Hutton Institute with a range of actives from different FRAG groups does not indicate any insensitivity issues with fungicide active ingredients other than fluazinam and phenylamides. However, it did show that EU_36_A2 and EU_37_A2 isolates formed consistently larger foliar lesions than those of the older lineages at very low dose rates of several key fungicide active ingredients. This supports aggressiveness testing conducted at INRAE, as part of the IPMBlight2.0 project that revealed that EU_36_A2 isolates tested formed larger average lesion sizes with abundant sporulation. Such properties are likely to make these lineages more difficult to manage and may explain the way they are displacing other clones across many European crops.
Lastly, the genetically diverse ‘Other’ samples comprised 26.2% of the sampled population in 2019, a frequency that has remained similar since 2014. These diverse types are always more prevalent in crops in the north and east of Europe and are consistent with a soil-borne source of oospores. Although there are epidemiological threats of earlier primary inoculum and theoretical evolutionary advantages to sexual recombination generating new pathogen phenotypes, we do not yet fully understand the practical threats posed by ‘Other’ strains of the pathogen compared to the clones. As part of the IPMBlight2.0 project the aggressiveness and virulence traits of a sample of these ‘Other’ isolates was tested, but did not highlight any clear trends.
The EuroBlight model of pathogen tracking is a rapid, cost-effective and co-ordinated approach to understanding pathogen evolution on a European scale. Data on the dominant clones have been passed to growers, advisors, breeders and agrochemical companies to provide practical management advice and shape longer-term strategies. The data provide an early warning of the incidence and spread of novel clones and the case of EU_37_A2 demonstrates such a timely response.
The EuroBlight network continues to harmonise methods with other networks in the Americas, Asia and Africa and encourages continued co-operation between groups involved in managing late blight to exploit the database and tools for improved awareness and blight management on a global scale. We will continue the project in 2020, so please contact the project team if you would like more information or if you would like to contribute. We thank all the partners who have contributed samples and supported the project.
Companies and institutions that participated in the sampling and sponsored the project (2013-2019)
Aarhus University, ACVNPT, ADAMA, AFBI, Agrifirm, Agricultural Institute of Slovenia, Agriphar, AHDB Potatoes, ARVALIS-Institut du Végétal, BASF SE, Bayer CropScience AG, Bayerische Landesanstalt für Landwirtschaft, Belchim Crop Protection, BSV Network (France), Centre Wallon de Recherches Agronomiques, Certis, Cheminova, Corteva CropSolutions, CUConsulting, CZAV, Delphi, Emsland Group, Estonian University of Life Sciences, Eurofins, Germicopa/ Florimond Deprez, Hochschule Osnabrück, HZPC Holland B.V., INRAE, Institute of Plant Protection and Environment in Serbia, The James Hutton Institute, Neiker, Nordisk Alkali, NIBIO, PCA, The Plant Breeding and Acclimatization Institute (IHAR), Profytodsd, Swedish University of Agricultural Sciences, Syngenta Agro GmbH, SynTech Research, Staphyt, TEAGASC, Technical University of Munich, and Wageningen University.
Contacts
Jens G. Hansen & Poul Lassen, Aarhus University. Contact: jensg.hansen@agro.au.dk
David Cooke & Alison Lees: James Hutton Institute, Dundee. Contact: david.cooke@hutton.ac.uk
Geert Kessel & Huub Schepers, Wageningen University and Research Centre. Contact: geert.kessel@wur.n
Didier Andrivon & Roselyne Corbiere, INRAE. Contact: Didier.Andrivon@inrae.fr
Download the news story as pdf
 |